StudentShare


Our website is a unique platform where students can share their papers in a matter of giving an example of the work to be done. If you find papers
matching your topic, you may use them only as an example of work. This is 100% legal. You may not submit downloaded papers as your own, that is cheating. Also you
should remember, that this work was alredy submitted once by a student who originally wrote it.
Login
Create an Account
The service is 100% legal
- Home
- Free Samples
- Premium Essays
- Editing Services
- Extra Tools
- Essay Writing Help
- About Us
✕
- Studentshare
- Subjects
- Health Sciences & Medicine
- Molecular Techniques in Cancer Research
Free
Molecular Techniques in Cancer Research - Assignment Example
Summary
The paper "Molecular Techniques in Cancer Research" highlights that using clinical applications, the abundant data accrued from the molecular profiling benchmark has encouraged a move away from the traditional broad therapeutic approach to cancer, towards a more developed strategy…
Download full paper File format: .doc, available for editing
GRAB THE BEST PAPER99% of users find it useful
- Subject: Health Sciences & Medicine
- Type: Assignment
- Level: Undergraduate
- Pages: 10 (2500 words)
- Downloads: 0
- Author: selinazboncak
Extract of sample "Molecular Techniques in Cancer Research"
Molecular Techniques in Cancer Research By Molecular Techniques in Cancer Research Question Approaches to Identify Targets of Phosphatase within Phosphoproteome
Protein phosphatases are important regulators for transducing signals. The actions for their cellular mechanisms are not well understood. Therefore, to identify their targets within phosphoproteome, a proteomics surveys is done. SILAC-based quantitative Mass Spectrometry to measure the differences in phosphorylation and protein expression upon the ablation of phosphatase in phosphoproteome is carried out. Fractionation of phosphopeptide using chromatography of strong cation exchange that is combined with IMAC (Immobilized Metal Affinity Chromatography) enrichment enables the quantification of sites for distinct phosphorylation in the wild type of Ppt1 versus the yeast cells of Ppt1-deficient. The quantification of 1897 yeast protein is done.
Here the outcome involves the detection or no detection of the main changes of protein that accompanies the deficiency of Ppt1. Additionally, one can find the number of phosphorylation sites and events repressed in the cells that lack Ppt1. Ppt1 tends to act on their targets in a sequential fashion. Most of the phosphoproteins need to be involved in responding to heat stress in accordance with Ppt1 functions. The results will show the quantitative and large-scale phosphoproteonics in identifying the targets of Phosphatase action within Phosphoproteome.
General Workflow and Experimental Design
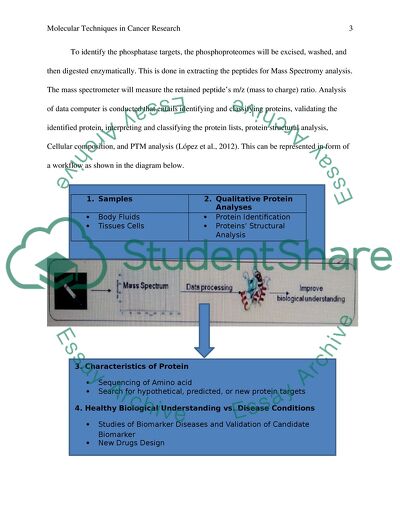
To identify the phosphatase targets, the phosphoproteomes will be excised, washed, and then digested enzymatically. This is done in extracting the peptides for Mass Spectromy analysis. The mass spectrometer will measure the retained peptide’s m/z (mass to charge) ratio. Analysis of data computer is conducted that entails identifying and classifying proteins, validating the identified protein, interpreting and classifying the protein lists, protein structural analysis, Cellular composition, and PTM analysis (López et al., 2012). This can be represented in form of a workflow as shown in the diagram below.
Advantages and Disadvantages of Strategies
Edman sequencing does not give provisions for high-throughput analysis and is an operation that is labour intensive. The advantage of incorporating the Mass Spectromy tool, is the ability of developing protein residues of phosphorylated. Conversely, the antibodies of phosphatase are used in immunoprecipitating and thus enriching the proteins from the complicated mixtures. The disadvantage of this is that, the commercial available antibodies are absent. Therefore, the proteins need to be enriched and purified from the complex mixtures. Additionally, conducting the proteins in solution digestion, the phosphopeptides can be loaded in various chromatographies (Hancock et al., 2008). The enriched solution can be presented into various reverse phase chromatography to desalt and clean the phosphopeptides. Moreover, the chromatographies minimize the phosphorylated peptides suppression in mass spectra. In IMAC strategy, the phosphopeptides that are negatively charged undergo purification by their ability to metal ions that are positively charged. However, some of the strategies are negatively affected by the problem of acidic binding and non-phosphorylated peptides. The disadvantage of this strategy is seen when the methyl esterification increase the complexity of spectra and needs the sample’s lyophilization thus causing the adsorptive losses in phosphopeptides (Albers et al., 2013).
Resources for Phosphate Identification
Some of the available resources that will enable one sort quickly through data during the process of phosphate identification include the GenBank and Mass Spectrometer. The GenBank uses TFASTF and FASTF programs (Damer, 1998). The technology sequences the proteins that were recovered from the affinity chromatography. Subsequently, the mass spectrometer with the help of computers is one of the resources that perform timely data sorting. The two components of the resource (gas chromatograph and mass spectrometer) allow substance identification with the help of the pure sample and traditional detector for mass spectrometry and gas chromatography respectively.
Question 2
Methods to Characterize “Interactome” of protein X
Interactomes are important in every cellular processes starting from the metabolism process to structural process. Elucidating the individual associations of protein their complex interaction network is an important objective of functional genomics. For instance, discovering the interactome for protein X can offer insight into the right operation beyond what the sequence-based predictions are and offer a benchmark for future research. The synthetic genetic methods like two-hybrid screening normally reveal the surprising array of interactomes for any targeted protein (“Overview of Protein-Protein Interaction Analysis”, 2013). This screening approach can produce false-positive outcomes, and hence putative interactors must be ascertained by independent techniques. In Vitro biochemical methods to characterize the interactome are time honored and varied, some being old. Therefore, the five methods for characterizing the interactome include the protein cross-linking, binding assays, rate-zonal centrifugation, co-immunoprecipitation, and blue native electrophoresis (Dey et al., 2012).
Co-Immunoprecipitation
Here, the target protein after precipitation is used in co-precipitating the protein complex from the lysate. The interactome is bound to the prey. The proteins under immunoprecipitation are detected by SDS-PAGE (Sodium Dodecyl Sulfate-polyacrylamide gel electrophoresis) and the analysis of western blot. Assumption here is that the protein has relationship with the target antigen.
Blue Native PAGE
Similar to Co-IP; since they use the beaded support in purifying the interacting proteins. They differ in the way they capture their protein complexes; Co-IP employs the antibodies while the pull-down assays uses bait. Pull-Down assays are important to study the interactions that are stable or strong (Alhosin et al., 2011).
Chemical Cross-Linking
Majority of the interactome are transient, since they occur briefly as one of the cascade or other functions of metabolism within a cell. The method tends to permanently or stabilize adjoin the interaction complex’ components. After the components have been cross-linked, electrophoresis, mass spectrometry, affinity purification, and cell analysis will be used in analyzing the interactome while maintaining the interacting complex.
In Vitro binding
This method involves crosslinking the prey and bait proteins with the help of a crosslinking agent that is labeled and cleaving the nexus between the interactomes so that preys remain attached to prey (Miernyk & Thelen, 2008). The method is important since it has the ability of identifying proteins that interact transiently and weakly with the interested protein. Upcoming non-isotopic reagents and techniques continue to make the technique more simple and accessible to carry out.
Rate Zonal Sedimentation
A blue Native PAGE assay is different from Co-IP in terms of their detection of interactomes using the tagged proteins rather than antibodies. For Rate Zonal Sedimentation, difference is seen with the Western Blot Analysis in the detection of the interactomes; they are detected using the incubating electrophoresed protein using a tagged and purified bait protein. The term far was generated to emphasize the distinction.
Methods and their Interactions
Method
Interactions
Co-Immunoprecipitation
Strong or Stable
Blue Native PAGE
Strong or Stable
Chemical Cross-Linking
Weak or Transient
In Vitro Binding
Weak or Transient
Rate Zonal Sedimentation
Moderately stable.
Relative Merits of the Methods
Method
Merits
Demerits
Rate Zonal Sedimentation
it is not destructive, compatible with various techniques of downstream analysis and highly reproducible
The method is time intensive and needs specialized tools and instruments.
Blue Native PAGE
The method is cheap, needs to specialized tool, compatible with various techniques if downstream analysis
it operates best with many membrane proteins, its resolution is low, and is very sensitive to the weight solutes
Co-IP
it is highly specific, simple, and compatible with various downstream analysis, and its reagents can be recycled
It has low capacity and experiences difficulties to obtain antibodies having high specificity
In vitro binding
It is simple and compatible with various downstream analysis methods
Manipulation of the DNA is necessary, and is unsuitable when a third party is mediating the interaction
Chemical Cross-Linking
it has a higher capacity and tends to stabilize specific interactions
The method complicates the downstream analysis. It has potential for the interference of the interactions.
Question 3
Role of Green Fluorescent Protein in Tumorigenesis
Fluorescent proteins have been used during cancer research cancer. The protein has been used to detect and track the specific cell lineages of cancer during the development of tumor. It has also been collaborated to protein to act with their localization. The experimental model of tumor process’ induction by applying the tumor cells enables the likelihood of studying various mechanisms (Hicks, 2012). The tumorigenesis opens up important information based on the complex interactions between the tumor tissues and the host organism. One of the crucial endeavors in the experiment is to comprehend how the various types of cells behave. Therefore, tumorigenesis visualizes the tumor cells found in vivo. However, various methods of studying the colonization, growth, and cancer dormancy need extensive sacrifice and historical preparation of the animal or tissue.
Optimal imaging with the help of fluorescence expression of Green Fluorescent Protein resolves the problems of real-time imaging techniques and enables the tumorigenesis in viable living animals or fresh tissue. Various groups have chosen the tumor cell line in stabilizing the express Green Fluorescent Proteins at higher level both in vivo and in vitro. The cells can undergo transplant to animals and can be detected for subsequent colonization in various organs (Chalfie & Kain, 2006). The main advantage of the Green Fluorescent Protein expressing humor is that the imaging does not require any procedure, substrates, contrast agents, light-tight boxes, or anesthesia. Tumor cells’ transfection with Green Fluorescent Protein produces a stable, heritable cytoplasmic marker that enables the cells to undergo detection for longer observations in vivo. In studying the tumor cells behaviors, the expression of Green Fluorescent Protein can be used in various tumor cell lines such as lung cancer, colon cancer, ovarian cancer, pancreatic cancer, and melanoma. The studies concur that tumor cells that are transfected with GFP genes are strong instruments in vivo visualization for growth of tumor, angiogenesis, and tumor microenvironment (“Protein–protein interaction screening”, (2012.). To find out if the tumor will be induced, is possible after a long period after the application of tumor cells.
The experimental strategy involved studying the alterations in brain section that process the sensory information rats that bear the tumor. The aim of the experiment was to come up with appropriate experimental model of the tumor that will help in studying the interaction of the peripheral tumor and the brain. It was found out that intraperitoneal application of the BP6 cells into the rats produced inductions of detectable tumor found in specified compartment of the organism. In the animal tumor, it was found out that the changes of endocrine and immune organs were present in the chosen sections of the brain. Therefore, BP6 is labeled by incorporating Green Fluorescent Protein ‘gene. The protein’s gene is integrated in the chromosome (Shin et al., 2012). The process of cell division will then pass the protein gene to the subsequent cells generation. According to studies, the protein transduction will not cause the growth inhibiting or cytopathology effects on the tumor cells, gene-encoding proteins do not have a bad effect on the evaluation of promotor activity. The approach is normally used in various cell lines of the tumor.
The hypothesis suggests the interference existing between the BP6 cells property and GFP. It looks that the cells of BP6 having incorporated GFP gene proliferate in vivo below BP6 in absence of GFP incorporated gene. On the GDP’s immune response and that, Green Fluorescent Protein acts as antigens (Liu et al., 2012). Due to significance of protein molecules, the importance of Green Fluorescent Protein cannot be repudiated. Due to progressive progress of researches, there is prevalence use of the Green Fluorescent Protein in marking the tumor in organisms. In summary, Green Fluorescent Protein reports the activation of Tcf4 in vivo and keeps records the predicted rise in the activation of Tcf4 that arises from the inactivation of Apc.
Question 4
Anti-body Based Techniques
To implement the effective anti-body based techniques in therapeutically regimes of cancer depends on the translation and identification of the informative biomarkers that help in clinical decision-making. Antibody-based techniques occupy an important space in the discovery of biomarker and validation pipeline. This tends to facilitate a high throughput monitoring of the candidate markers (Gilliam, 2007). Although the utility of the emerging technology has not yet been established, the conventional use of the antibodies as one of the affinity reagent in the predictive assays and clinical diagnostics suggest that the first translation of the approached is a goal that can be achieved. Additionally, in liaison with the trascriptomic and genomic methods to stratify patients, the antibody-based approaches provide the promise of insights into states with cancer disease.
Personalized medicine needs the application and discovery of unambiguous predictive, prognostics, and pharmacodynamics biomarkers in informing the therapeutic decisions (Disis, 2006). Screening methods that are high-throughput especially the transcriptomic and genomic profiling have improved the knowledge of tumorigenesis molecular basis, therapeutic response, and disease progression. Therefore, the regimen of individualized treatment is seen to be a goal that can be achieved. The role of the antibodies is incorporate development of predictive biomarkers as highlighted by success to detect the estrogen receptor and ERBB2 expression found in breast cancer. In eras of post genomic, the development progress of assays that are clinically implemented has not maintained the pace with the biomarker recovery rate. As a result, a pressing concern exists for innovative and improved strategies to expedite the cancer biomarkers translation into the arena of clinics (Adams & Sheehan, 2013). The antibody-based techniques offer a logical technique for systematic generation and using the specific antibodies for the exploration of proteome.
Through enabling technologies, the regime and using antibodies for profiling of proteins on a global arena is a proper approach that allows the systematic exploration of human protein using various high-throughput assays that include IHC (Including Immunohistochemistry ) on TMAs ) Tissue Microarrays, and pathway analysis with the help of RPPAs (Reverse Phase Protein Arrays) and diagnostic assays that are serum-based. Founded based in their ability to validate and systematically generate sensitive and specific antibodies, the versatile assays are on the verge of generating the predictive assays and molecular diagnostics that are needed in facilitating the cancer therapy’ personalization (Khleif, 2008).
Using clinical applications, the abundant data accrued from the molecular profiling benchmark have encouraged a move away from the traditional broad theraupeutic approach to cancer, towardsa more developed strategy. The growth of personalized protocols are dependent in the growth and development of the well validated, robust, pharmacodynamics assays and informative predictive.ERRB2 and ER were crucial as early illustrations of the predictive biomarkers and epitomize personalized in breast cancer. Recently, approaches that are transcriptomic have resulted in the advancement to therapeutically protocols. Another technique is translating the assays to clinic (Brennan et al., 2010). Despite the massive advancement in the preclinical setting, the new assays translation to the clinic has recorded a low figure. The problem tends to be multifactorial; however, various issues have been brought forward over the last years. It is therefore accepted that predictive markers will benefit from the personalized therapeutic regime. This posed to be a considerable predicament for solid tissue.
Reference
1. Adams, V. R., & Sheehan, J. B. 2013. Guide to cancer chemotherapeutic regimes. New York: McMahon Publishing Group.
2. Albers, H. M., Ovaa, H., Bakker, J., Kuijl, C., Neefjes, J., Celie, P., et al. 2013. Integrating Chemical and Genetic Silencing Strategies To Identify Host Kinase-Phosphatase Inhibitor Networks That Control Bacterial Infection. ACS chemical biology, 102, 131125093526007.
3. Alhosin, M., Sharif, T., Mousli, M., Etienne-Selloum, N., Fuhrmann, G., Schini-Kerth, V. B., et al. 2011. Down-regulation of UHRF1, associated with re-expression of tumor suppressor genes, is a common feature of natural compounds exhibiting anti-cancer properties. Journal of Experimental & Clinical Cancer Research , 301, 41.
4. Berggård, T., Linse, S., & James, P. 2007. Methods for the detection and analysis of protein–protein interactions. PROTEOMICS, 716, 2833-2842.
5. Brennan, D. J., Oconnor, D. P., Rexhepaj, E., Ponten, F., & Gallagher, W. M. 2010. Antibody-based proteomics: fast-tracking molecular diagnostics in oncology. Nature Reviews Cancer, 109, 605-617.
6. Chalfie, M., & Kain, S. 2006. Green fluorescent protein: properties, applications, and protocols 2nd Ed. Hoboken, N.J.: Wiley-Interscience.
7. Damer, C. K. 1998. Rapid Identification of Protein Phosphatase 1-binding Proteins by Mixed Peptide Sequencing and Data Base Searching. CHARACTERIZATION OF A NOVEL HOLOENZYMIC FORM OF PROTEIN PHOSPHATASE 1. Journal of Biological Chemistry, 27338, 24396-24405.
8. Dey, B., Thukral, S., Krishnan, S., Chakrobarty, M., Gupta, S., Manghani, C., et al. 2012. DNA–protein interactions: methods for detection and analysis. Molecular and Cellular Biochemistry, 3651-2, 279-299.
9. Disis, M. L. 2006. Immunotherapy of cancer. Totowa, N.J.: Humana Press.
10. Gilliam, L. K. 2007. An Immunotherapeutic Approach to the Treatment and Prevention of Breast Cancer, Based on Epidermal Growth Factor Receptor Variant, Type III. Ft. Belvoir: Defense Technical Information Center.
11. Hancock, J. T., Henson, D., Nyirenda, M., Desikan, R., Harrison, J., Lewis, M., et al. 2008. Proteomic identification of glyceraldehyde 3-phosphate dehydrogenase as an inhibitory target of hydrogen peroxide in Arabidopsis. Plant Physiology and Biochemistry, 439, 828-835.
12. Hicks, B. W. 2012. Green fluorescent protein: applications and protocols. Totowa, NJ: Humana Press.
13. Khleif, S. 2008. Tumor immunology and cancer vaccines. Boston: Kluwer.
14. Liu, F., Frangioni, J. V., Henske, E. P., Choi, H. S., Yu, J., Gibbs, S. L., et al. 2012. Real-Time Monitoring of Tumorigenesis, Dissemination, & Drug Response in a Preclinical Model of Lymphangioleiomyomatosis/Tuberous Sclerosis Complex. PLoS ONE, 76, e38589.
15. López, E., Wang, X., Madero, L., López-Pascual, J., & Latterich, M. 2012. Functional phosphoproteomic mass spectrometry-based approaches. Clinical and Translational Medicine, 11, 20.
16. Miernyk, J. A., & Thelen, J. J. 2008. Biochemical approaches for discovering protein-protein interactions. The Plant Journal, 534, 597-609.
17. Overview of Protein-Protein Interaction Analysis. n.d.. Overview of Protein-Protein Interaction Analysis. Retrieved June 30, 2014, from http://www.piercenet.com/method/overview-protein-protein-interaction-analysis
18. Protein–protein interaction screening | Online references | cyclopaedia.net. n.d.. Protein–protein interaction screening | Online references | cyclopaedia.net. Retrieved June 30, 2014, from http://www.cyclopaedia.info/wiki/Protein-protein-interaction-screening
19. Schreiber, T. B., Mäusbacher, N., Soroka, J., Wandinger, S., Buchner, J., & Daub, H. 2012. Global Analysis of Phosphoproteome Regulation by the Ser/Thr Phosphatase Ppt1 in Saccharomyces cerevisiae. Journal of proteome research, 114, 120227101538005.
20. Shin, J., Epstein, J. A., He, S., Guo, F., Dahlberg, S., Haidar, S., et al. 2012. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Disease models & mechanisms, 56, 881-894.
Read
More
CHECK THESE SAMPLES OF Molecular Techniques in Cancer Research
Molecular biology cloning and Brachyury gene
Brachyury gene: definition and the relation between the genes with cancer Brachyury refers to a protein which is encoded by the T gene in humans.... Brachyury also helps in defining the mesoderm in the process of gastrulation (National cancer institute, nd, para 5).... Genetic evaluations of individuals with hereditary form of bone cancer reveal that acquiring an additional T gene might cause the uncommon bone cancer.... A comprehensive hereditary examination revealed that chromosome 6 section was more apt to be related with hereditary bone cancer....
4 Pages
(1000 words)
Essay
The Effect of Oxaliplatin Genotoxicity on Human Lymphocytes
This has been approved for treatment of metastatic colon cancer in combination with 5-fluorouracil and folinic acid8.... Its preclinical activity against colorectal cancer has been studied in great detail.... It has been suggested that oxaliplatin has a greater extent of cell kill in resistant cancers since therapy with this agent may result in greater resistance to repair mechanisms leading to affected recovery of cancer cells11.... In this assignment contemporary literature will be reviewed to address the effects of oxaliplatin genotoxicity on human lymphocytes by using various cytogenetic techniques....
25 Pages
(6250 words)
Essay
Review of the Maldi-MS Technique
The paper "Review of the Maldi-MS Technique" discusses that flow cytometry is more sensitive in the detection of some diseases.... Bone marrow biopsy and flow cytometry were used to detect minimal residual disease (MRD) in patients in remission for chronic lymphocytic leukemia.... .... ... ... Mass spectrometry (MS) is a technique used to measure the mass-to-charge ratio of particles (Jensen, Shevchenko, & Mann, 1997)....
8 Pages
(2000 words)
Coursework
Drugs that Bind DNA to Treat Diseases
Nucleic Acids research, 33 (18), 6034-6047.... cancer Informatics, 6, 315-355.... he three articles were selected because they show a wide range of techniques and results of experiments that explained how different anti-cancer drugs interact with DNA and DNA structure.... Their findings show different ways in how cancer can be attacked, and how future drugs can be designed to be more effective....
6 Pages
(1500 words)
Case Study
Proteomic Approaches and Reviews on Cancer Biomarker Researches
This work "Proteomic Approaches and Reviews on cancer Biomarker Researches" describes the war against cancer.... Biomarkers are among the devised way of fighting cancer by detecting it early and in the prevention of cancer along with enhancing their treatment.... A cancer biomarker refers to a biological molecule found in the body tissues, fluids, and the blood that are an indication of a normal or abnormal process of cancer....
8 Pages
(2000 words)
Literature review
Way of Identifying Genes that Have Developed Resistance to Targeted Therapies in Molecular Cancer
While microarray techniques represent a fundamental opportunity for clinical research, significant issues must be considered.... "Way of Identifying Genes that Have Developed Resistance to Targeted Therapies in Molecular cancer" paper examines two approaches that are used to identify genes that have roles in drug resistance including microarray and next-generation RNA sequences.... These amounts Traditional approaches sequences are wildly used as guide therapy to treat patients with cancer....
10 Pages
(2500 words)
Assignment
Transcriptomics as a Powerful Tool in Understanding Gene Expression
Transcriptomics is the study of the transcriptome; it is the section in molecular biology that involves the study of the messenger RNA molecules, originating from an individual or population of a particular cell type.... "Transcriptomics as a Powerful Tool in Understanding Gene Expression" paper argues that transcriptomics has done medical good, therefore universities and medical training facilities should take with high regard this field by availing sufficient resources to teach students and equip researchers....
9 Pages
(2250 words)
Essay
The Liquid Biopsies and Traditional Histopathological Approaches
On the other hand, the traditional biopsies involve tissue biopsies and can also be used in the diagnosis of cancer and other diseases, monitoring response to diseases, as well as, monitoring for residual infections2.... recision therapies that target specific genes are set to transform the way cancer patients receive care and treatment.... This also limits their ability to diagnose and monitor cancer.... The conventional methods of acquiring biopsy materials limit the ability of clinicians to establish the molecular status of their patients....
12 Pages
(3000 words)
Report
sponsored ads
Save Your Time for More Important Things
Let us write or edit the assignment on your topic
"Molecular Techniques in Cancer Research"
with a personal 20% discount.
GRAB THE BEST PAPER

✕
- TERMS & CONDITIONS
- PRIVACY POLICY
- COOKIES POLICY